This package makes it easy to visualize, manipulate, and export phylogenetic trees in R using an interactive viewer/editor. You can insert this tree viewer into Rmd/Quarto documents make your trees accessible online. If you want to use this for web development outside of R, consider the javascript heat-tree package which this package builds upon.
Quick start
The package includes example data sets that are automatically loaded with the package (example_tree_1), so you can try it out with minimal effort. After installing the packages, simply run the lines below to get an idea of how it works:
library(heattree)
heat_tree(example_tree_1, metadata = example_metadata_1, aesthetics = c(tipLabelColor = 'source'), layout = 'circular')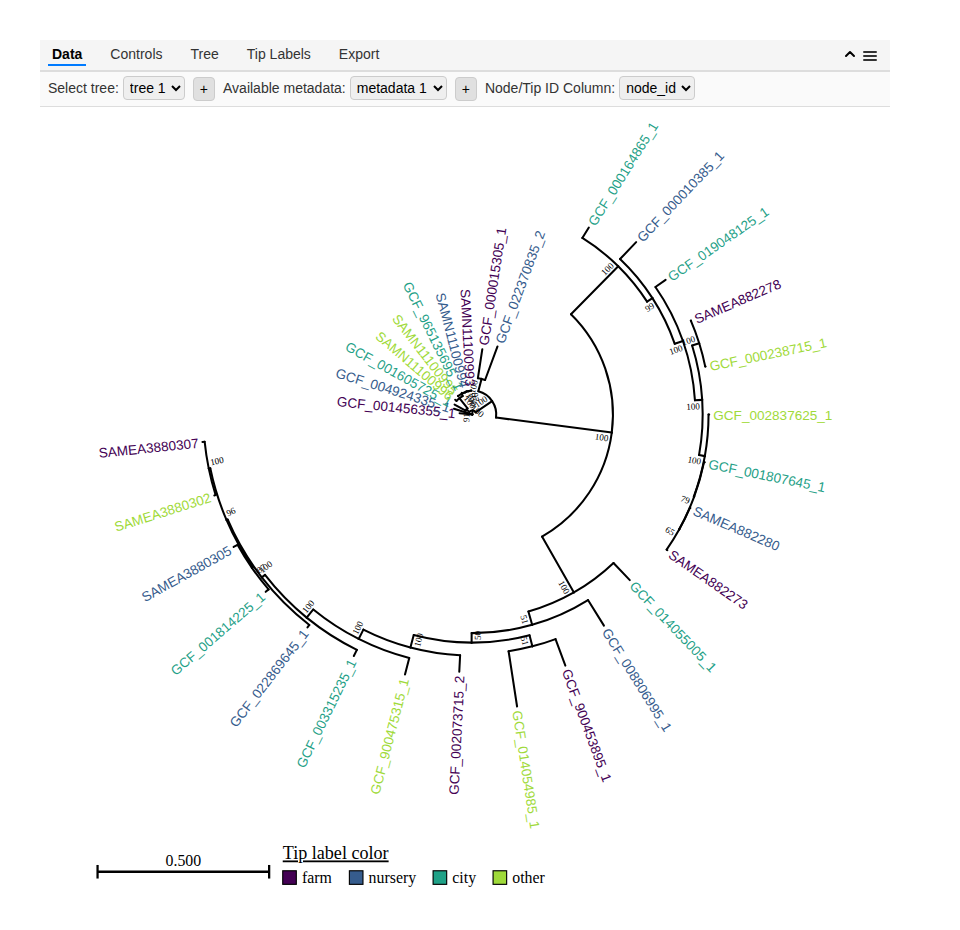
Contributing and feedback
Contributions and feedback are welcome! Please visit the GitHub repository to report issues or submit pull requests.